Note
Go to the end to download the full example code.
Gaussian Process Outlier Detection
Using Gaussian Processes (GPs) to detect outliers/anomalies in data, with per-data-point noise variances. Plotting final results with color-coded error bars to indicate each point’s learned noise level.
import torch
import torch.optim as optim
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import matplotlib.colors as mcolors
torch.manual_seed(0)
<torch._C.Generator object at 0x7fb997745290>
Create synthetic data with discontinuity
N = 100
min_data, max_data = -20, 20
x_train = torch.linspace(min_data, max_data, N).reshape(-1, 1)
f_true = 2 * torch.sigmoid(x_train) - 1
noise = 0.5 * torch.randn_like(f_true) / (f_true.abs() ** 5 + 1e-1)
y_train = f_true + noise
Setup GP
Define squared-exponential kernel
def rbf_kernel(X1, X2, lengthscale, variance):
"""
k(x1, x2) = variance * exp(-(x1 - x2) ^ 2 / (2 * lengthscale ^ 2))
"""
sqdist = (X1 - X2.T).pow(2)
return variance * torch.exp(-0.5 * sqdist / lengthscale.pow(2))
Negative log marginal likelihood
def negative_log_marginal_likelihood(x, y, lengthscale, variance, noise_weights):
"""
-log p(y|x) = 0.5*y^T(K^-1)*y + 0.5*log|K| + 0.5*N*log(2*pi),
with K = RBF + diag(noise_weights).
"""
K = rbf_kernel(x, x, lengthscale, variance)
K = K + torch.diag(noise_weights)
L = torch.linalg.cholesky(K)
alpha = torch.cholesky_solve(y, L)
log_det_K = 2.0 * torch.sum(torch.log(torch.diagonal(L)))
N_data = x.shape[0]
nll = 0.5 * y.T.mm(alpha) \
+ 0.5 * log_det_K \
+ 0.5 * N_data * torch.log(torch.tensor(2.0 * 3.141592653589793))
return nll.squeeze()
Posterior prediction for new test points
def gp_posterior(x_train, y_train, x_test, lengthscale, variance, noise_weights):
"""
Returns (mean, covariance) at x_test for GP with RBF kernel + per-data-point noise.
"""
K_xx = rbf_kernel(x_train, x_train, lengthscale, variance)
K_xx = K_xx + torch.diag(noise_weights)
L = torch.linalg.cholesky(K_xx)
K_xs = rbf_kernel(x_test, x_train, lengthscale, variance)
K_ss = rbf_kernel(x_test, x_test, lengthscale, variance)
alpha = torch.cholesky_solve(y_train, L)
pred_mean = K_xs.mm(alpha)
v = torch.linalg.solve(L, K_xs.T)
pred_cov = K_ss - v.T.mm(v)
return pred_mean, pred_cov
Train model
def train(
n_steps=600,
learn_weights=True
):
lengthscale = torch.nn.Parameter(torch.tensor(1.0))
variance = torch.nn.Parameter(torch.tensor(1.0))
log_noise_weights = torch.nn.Parameter(torch.zeros(N) - 2.0) # log-variance, init small
params = [lengthscale, variance]
if learn_weights:
params.append(log_noise_weights)
optimizer = optim.Adam(params, lr=0.03)
nll_history = []
for step in range(n_steps):
optimizer.zero_grad()
noise_weights = torch.exp(log_noise_weights) # ensure positivity
nll = negative_log_marginal_likelihood(
x_train, y_train, lengthscale, variance, noise_weights
)
nll += noise_weights.sum() # L1 penalty on noise weights
nll.backward()
optimizer.step()
nll_history.append(nll.item())
if (step + 1) % 50 == 0 or step == 0:
print(
f"Step {step + 1}/{n_steps}, NLL = {nll.item():.4f}, "
f"lengthscale = {lengthscale.item():.4f}, "
f"variance = {variance.item():.4f}, "
f"noise (mean) = {noise_weights.mean().item():.4f}"
)
return nll_history, lengthscale, variance, log_noise_weights
Plot
def plot(nll_history, lengthscale, variance, log_noise_weights):
# Plot final GP fit (mean + 2 std)
# showing training points with error bars colored by noise level
x_test = torch.linspace(min_data, max_data, 200).reshape(-1, 1)
with torch.no_grad():
noise_weights = torch.exp(log_noise_weights)
pred_mean, pred_cov = gp_posterior(
x_train, y_train, x_test, lengthscale, variance, noise_weights
)
pred_std = torch.sqrt(torch.diagonal(pred_cov))
# Convert to numpy for plotting
x_train_np = x_train.squeeze().numpy()
y_train_np = y_train.squeeze().numpy()
noise_std_np = torch.sqrt(noise_weights).detach().numpy()
x_test_np = x_test.squeeze().numpy()
pred_mean_np = pred_mean.squeeze().numpy()
pred_std_np = pred_std.numpy()
plt.figure(figsize=(10, 10))
# Plot GP predictive mean +/- 2 std
plt.plot(x_test_np, pred_mean_np, 'r', label='GP Mean')
plt.fill_between(
x_test_np,
pred_mean_np - 2 * pred_std_np,
pred_mean_np + 2 * pred_std_np,
color='r', alpha=0.2, label='±2 Std. Dev.'
)
# Color-coded error bars for each training point
norm = mcolors.Normalize(vmin=noise_std_np.min(), vmax=noise_std_np.max())
cmap = cm.get_cmap('viridis')
# Plot each data point with vertical error bars according to noise_std
for i in range(len(x_train_np)):
c = cmap(norm(noise_std_np[i]))
plt.errorbar(
x_train_np[i], y_train_np[i],
yerr=noise_std_np[i],
fmt='o', color=c, ecolor=c, capsize=3
)
# Add a colorbar to show how noise std maps to color
sm = cm.ScalarMappable(norm=norm, cmap=cmap)
sm.set_array([]) # needed in some older matplotlib versions
cbar = plt.colorbar(sm, ax=plt.gca())
cbar.set_label('Learned Noise Std. Dev.')
plt.title('Gaussian Process Regression (RBF Kernel + Per-Point Noise)')
plt.xlabel('x')
plt.ylabel('y')
plt.legend()
plt.tight_layout()
plt.show()
# Plot training progress (NLL)
plt.figure(figsize=(8, 4))
plt.plot(nll_history, label='NLL')
plt.xlabel('Iteration')
plt.ylabel('Negative Log-Likelihood')
plt.title('Training Progress')
plt.legend()
plt.tight_layout()
plt.show()
print("\nLearned parameters after training:")
print(f" lengthscale = {lengthscale.item():.4f}")
print(f" variance = {variance.item():.4f}")
print(f" Mean noise weight = {noise_weights.mean().item():.4f}")
Run model
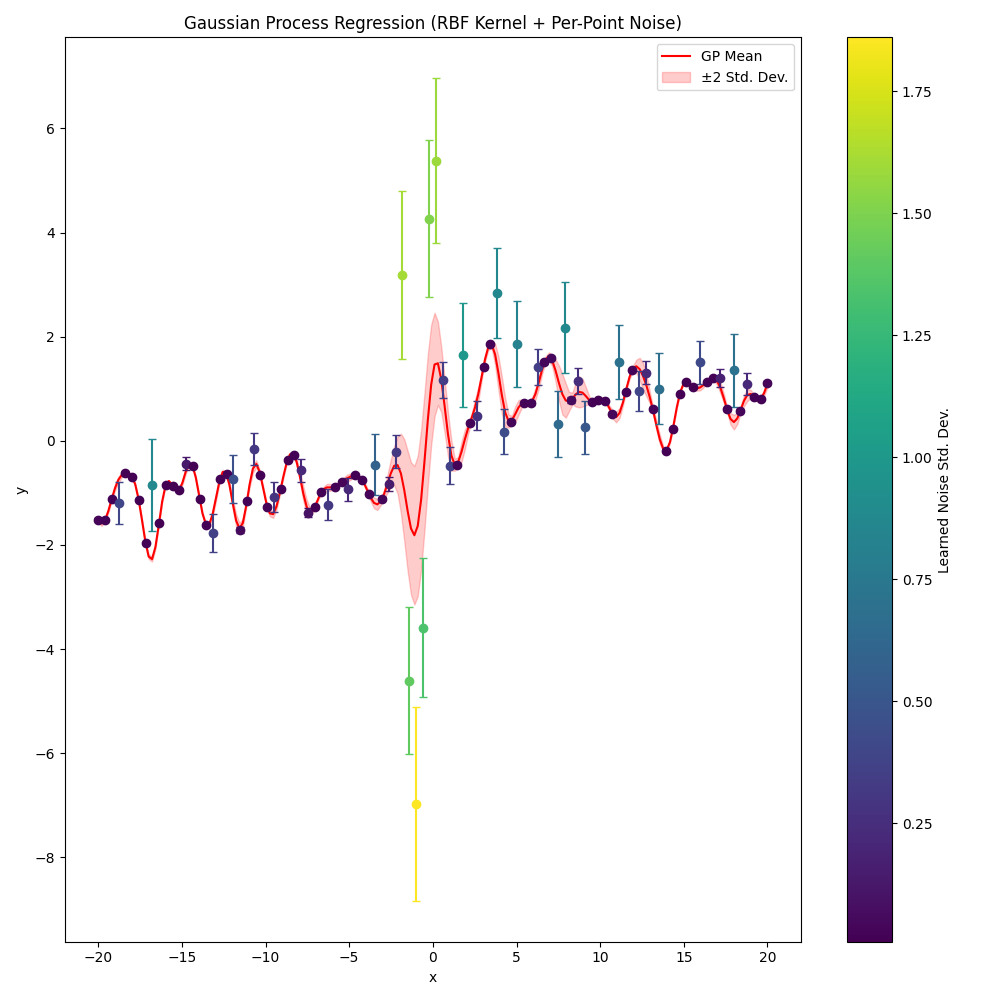
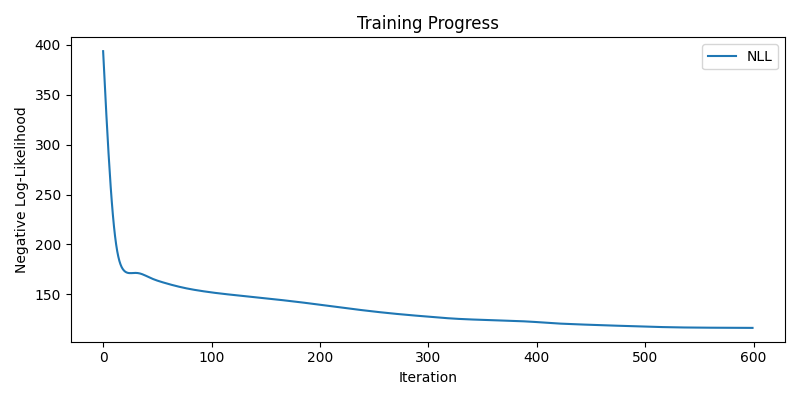
With learning weights
nll_history, lengthscale, variance, log_noise_weights = train(learn_weights=True)
plot(nll_history, lengthscale, variance, log_noise_weights)
Step 1/600, NLL = 393.4452, lengthscale = 0.9700, variance = 1.0300, noise (mean) = 0.1353
Step 50/600, NLL = 164.2169, lengthscale = 0.4453, variance = 1.7800, noise (mean) = 0.0976
Step 100/600, NLL = 152.1879, lengthscale = 0.5481, variance = 1.9608, noise (mean) = 0.1114
Step 150/600, NLL = 146.2200, lengthscale = 0.5849, variance = 2.0953, noise (mean) = 0.1152
Step 200/600, NLL = 139.8615, lengthscale = 0.6172, variance = 2.1408, noise (mean) = 0.1182
Step 250/600, NLL = 133.0010, lengthscale = 0.6438, variance = 2.0798, noise (mean) = 0.1309
Step 300/600, NLL = 127.8574, lengthscale = 0.6669, variance = 1.8822, noise (mean) = 0.1552
Step 350/600, NLL = 124.5856, lengthscale = 0.6902, variance = 1.6446, noise (mean) = 0.1774
Step 400/600, NLL = 122.4225, lengthscale = 0.7060, variance = 1.4754, noise (mean) = 0.1884
Step 450/600, NLL = 119.6359, lengthscale = 0.7339, variance = 1.3657, noise (mean) = 0.2046
Step 500/600, NLL = 117.8596, lengthscale = 0.7424, variance = 1.1870, noise (mean) = 0.2219
Step 550/600, NLL = 116.8072, lengthscale = 0.7464, variance = 1.0759, noise (mean) = 0.2315
Step 600/600, NLL = 116.5449, lengthscale = 0.7481, variance = 1.0636, noise (mean) = 0.2325
/home/runner/work/snippets/snippets/examples/plot_gaussian_proces_outlier_detection.py:167: MatplotlibDeprecationWarning:
The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
Learned parameters after training:
lengthscale = 0.7481
variance = 1.0636
Mean noise weight = 0.2325
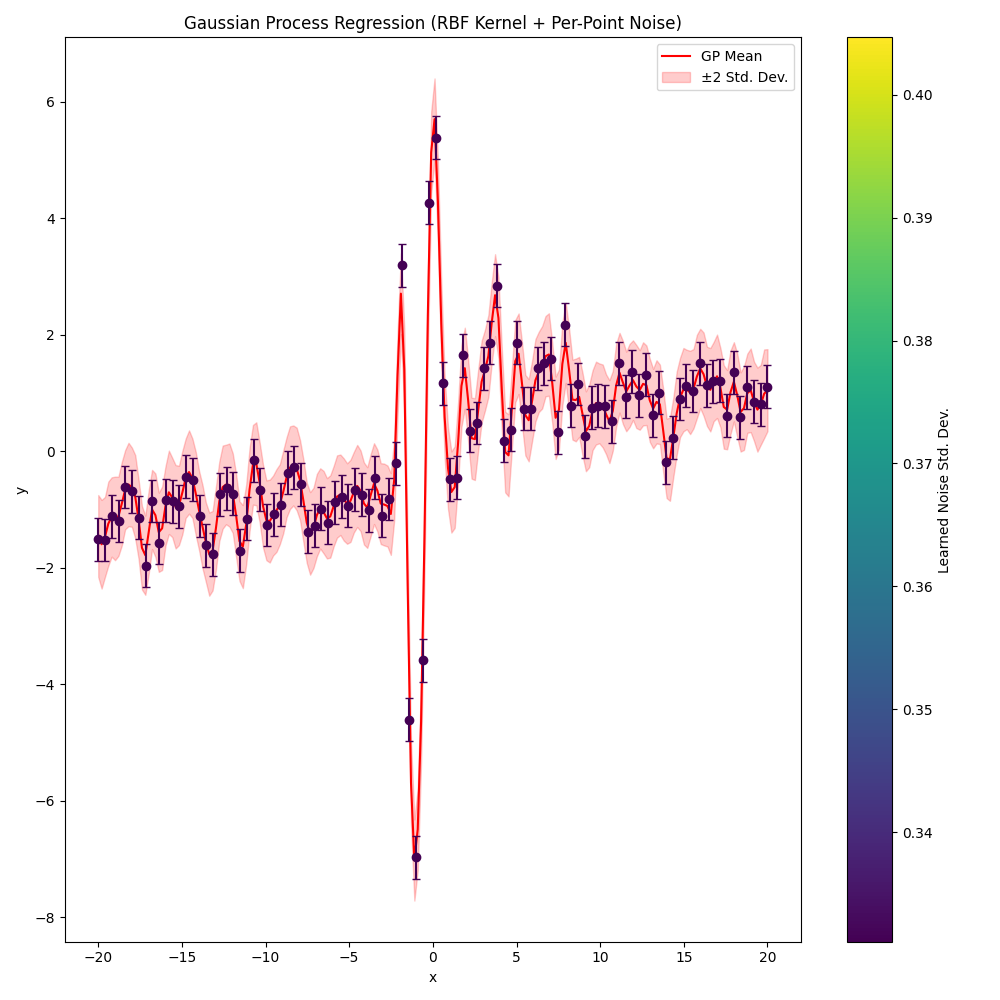
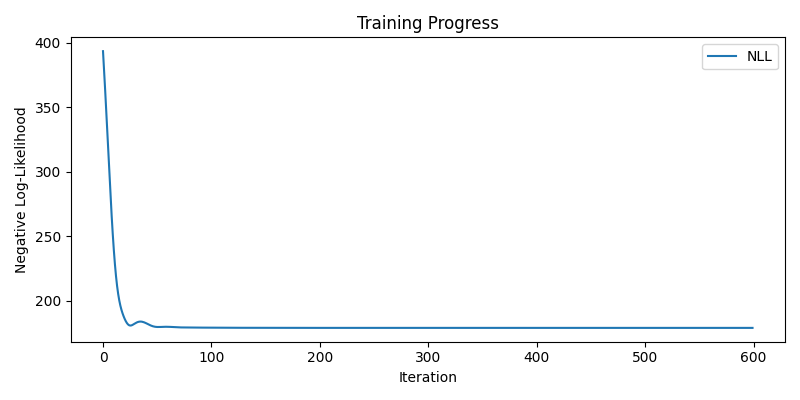
Without learning weights
nll_history, lengthscale, variance, log_noise_weights = train(learn_weights=False)
plot(nll_history, lengthscale, variance, log_noise_weights)
Step 1/600, NLL = 393.4452, lengthscale = 0.9700, variance = 1.0300, noise (mean) = 0.1353
Step 50/600, NLL = 179.6105, lengthscale = 0.3664, variance = 1.8482, noise (mean) = 0.1353
Step 100/600, NLL = 179.0276, lengthscale = 0.3769, variance = 2.0476, noise (mean) = 0.1353
Step 150/600, NLL = 178.8953, lengthscale = 0.3774, variance = 2.1564, noise (mean) = 0.1353
Step 200/600, NLL = 178.8651, lengthscale = 0.3791, variance = 2.2139, noise (mean) = 0.1353
Step 250/600, NLL = 178.8588, lengthscale = 0.3799, variance = 2.2421, noise (mean) = 0.1353
Step 300/600, NLL = 178.8577, lengthscale = 0.3802, variance = 2.2546, noise (mean) = 0.1353
Step 350/600, NLL = 178.8576, lengthscale = 0.3803, variance = 2.2595, noise (mean) = 0.1353
Step 400/600, NLL = 178.8575, lengthscale = 0.3804, variance = 2.2612, noise (mean) = 0.1353
Step 450/600, NLL = 178.8575, lengthscale = 0.3804, variance = 2.2617, noise (mean) = 0.1353
Step 500/600, NLL = 178.8575, lengthscale = 0.3804, variance = 2.2619, noise (mean) = 0.1353
Step 550/600, NLL = 178.8575, lengthscale = 0.3804, variance = 2.2619, noise (mean) = 0.1353
Step 600/600, NLL = 178.8575, lengthscale = 0.3804, variance = 2.2619, noise (mean) = 0.1353
Learned parameters after training:
lengthscale = 0.3804
variance = 2.2619
Mean noise weight = 0.1353
Total running time of the script: (0 minutes 2.518 seconds)